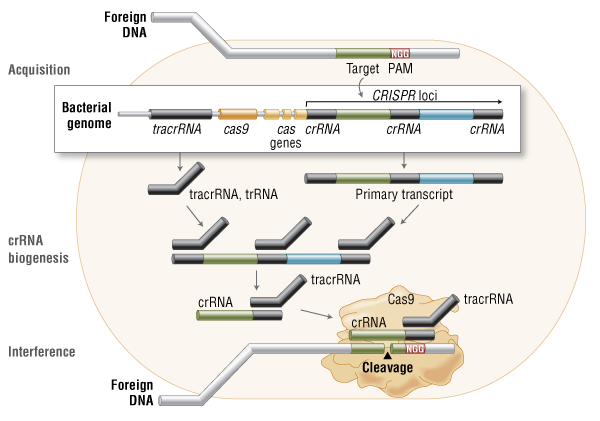
We human beings have dominated this world, but the world doesn't belong to human beings alone. There are many more species which not only help in maintaining the ecological and biological balance in the nature but are also useful to mankind in many ways. For example milk which is an important food all over the world, comes from animals. For meat and poultry, which again is consumed by a large number of people, has to come from animals and birds. Medicinal items and nutritional supplements are drawn both from natural and animal resources. We also keep animals as pet. A large population all over the world earns their living through farm animals including buffaloes, cows and goats. Though modern methods of cultivation have come, bullocks are still used for the purpose. All in all, animals very much belong to our world and we can't imagine our existence without them, in one way or the other.
Courses like MBBS, BDS etc. are much sought after by students completing their 10+2 with science subjects including biology. Such courses come under medical careers. People with these qualifications treat humans for different ailments. Animals and birds may also need treatment and there is a specialised field for that named as veterinary science. A person qualified in veterinary science is called veterinarian or vet (in short). The job of a veterinarian, basically, is to attend to the medical and healthcare needs of animals which include pets, livestock, zoo and laboratory animals. In cities and urban areas the most common animals to be treated by them are dogs and cats. In others they may have to attend to cows, bullocks, goats, sheep etc. Different segment of veterinarians may be involved in different requirements related to animals. Generally the job of a veterinarian consists of conducting physical examination, administering immunization, providing emergency and critical care, performing surgery and dental procedures and offering advice to people regarding care of their pets and livestock etc. Many of the animals maladies and infections may have adverse effect on people coming in the contact of such animals. So, veterinarians' role is also crucial to keep the human society safe and healthy. By monitoring and protecting the health of animals that provide food for a large section of population, veterinarians ensure a safe food supply for them. If you are not averse to animals, are comfortable in dealing with them and have aptitude for medical science, you may plan to grow as a veterinarian.
Subjects of study
As a student of veterinary science you will be exposed to both theoretical and practical aspects related to animal health, anatomy, physiology, biochemistry, pharmacology, parasitology, toxicology, epidemiology, genetics & breeding, nutrition, gynaecology and obstetrics, radiology, surgery, clinical and farm practices, livestock products technology and production management, veterinary clinical practices, livestock farm practices etc. Largely the course is divided into basic and clinical sciences.
Requirements to become a veterinarian
Formal education to become a veterinarian can be pursued at graduation level. The nomenclature for this graduation qualification is Bachelors in Veterinary Science, commonly known as B.V.Sc. In many cases the degree also includes the term animal husbandry. The entry requirements are same as applicable for MBBS. The applicant should have completed H.S.C. (i.e. 10+2) with physics, chemistry and biology or biotechnology with English. Selection is usually based on performance in entrance examination conducted for the purpose.
Various states conduct this entrance test for admission to veterinary institutes in their jurisdiction. While most of the states have a separate examination for entry to B.V.Sc., few have a common/ combined entrance examination for this and other professional courses like B.Tech., B.Pharm. etc.
Veterinary Council of India conducts a national All India Pre Veterinary entrance examination/test. 15 percent seats in recognised institutes across the country are filled on the basis of qualifying scores in this examination. There are nearly 40 such institutes where this quota of 15 per cent is applicable.
Entrance examinations are generally conducted with a duration of 2 to 3 hours in which objective questions from physics, chemistry, botany and zoology are asked.
The course duration for a degree in veterinary science is 5 and a half years of which one year is for intense internship. A B.V.Sc. degree opens the door to work as a vet. However many students prefer to join a master's degree course, M.V.Sc. after graduation. The course period for this postgraduate qualification is 2 years, divided into 4 semesters and eligibility criteria is B.V.Sc. pass. To be eligible for M.V.Sc. , candidates have to appear in the qualifying test conducted by Indian Council for Agricultural Research in collaboration with National Testing Agency. Doctoral (Ph.D.) opportunities are also available for those interested in research. University Grants Commission norms apply here as in case of Ph.D. in other subjects.
Selecting an Institute for study
Veterinary education is an old discipline of study offered at government universities and colleges since decades. Now few private institutes have also started courses in the subject.
Indian Veterinary Research Institutes (deemed to be university) at Izat Nagar, Bareilly is a premier establishment in veterinary education. However it offers course from postgraduation onwards only. Government has established a number of animal sciences universities in the country as given below:
Bihar Animal Sciences University,Patna
Nanaji Deshmukh Pashu Chikitsa Vigyan Vishwavidyalaya, Jabalpur
Sri Venkateswara Veterinary University, Tirupati
Lala Lajpat Rai University of Veterinary & Animal Science, Hisar, Haryana
U P Pandit Deendayal Upadhyay Pashu Chikitsa Vigyan Vishwavidyalaya evam Gau Anusandhan Sanstha, Mathura
Tamilnadu Veterinary & Animal Science University, Chennai
Chattisgarh Kamdhenu Vishwavidyalaya, Durg
West Bengal University of Animal & Fisheries Science, Kolkata
Karnataka Veterinary, Fisheries & Animal Science University, Bidar
Guru Angad Dev Veterinary & Animal Science University, Ludhiana
Maharashtra Animal & Fisheries Science University, Nagpur
Kerala Veterinary & Animal Science University, Wayanad
Kamdhenu University, Gandhinagar
Madhya Pradesh Pashu Chikitsa Vigyan Vishwavidyalaya, Jabalpur
Rajasthan University of Veterinary & Animal Science, Bikaner
Prof Jayashankar Telangana State Agricultural Veterinary University, Hyderabad
Other than above there are a number of colleges/universities which offer graduate/postgraduate courses in veterinary science and animal husbandry, some of which are listed below. where you may pursue a degree. The list is not in any particular order and is for indicative purpose only:
Dr. Rajendra Prasad Agricultural University, Pusa
Birsa Agricultural University, Kanke, Ranchi
Assam Agricultural University, Jorhat
Indira Gandhi Agricultural University, Raipur
Anand Agricultural University, Anand, Gujarat
Govind Ballabh Pant University of Agriculture & Technology, Pantnagar
Junagadh Agricultural University, Gujarat
P.V. Narsimha Rao Telangana Veterinary University, Hyderabad
Sardarkrushinagar Dantwada Agricultural University, Banskantha, Gujarat
Chaudhary Sarvan Kumar Himachal Pradesh Krishi Vishwavidyalaya, Palanpur
Swami Keshvanand Rajasthan Agricultural University, Bikaner
Narendra Dev University of Agriculture & Technology, Faizabad
Pondicheri University
Orissa University of Agriculture & Technology, Bhubaneshwar
Central Agricultural University ,Imphal
Jawahar Lal Nehru Krishi Vishwavidyalaya, Jabalpur
College of Veterinary Science & Animal Husbandry, Salasih, Mizoram
Work opportunities
Supply of Veterinary professionals is limited so it may be relatively easier to find a job or settle down in career. This position shouldn't be interpreted as lack of competition in job market .So you need to be better equipped. Those interested to work in government may join government veterinary hospitals at district and taluka levels which exist parallel to government general hospitals/primary health centres. In many states District Livestock Development Officers (DLOs) are appointed who in many cases are considered to be the incharge of Veterinary hospitals.
One good opportunity for which only a graduate qualification is needed comes from public sector banks who allow candidates with B.V.Sc. to join as Agricultural Field Officer/Rural Development officer. Selection is based on written test and interview. Private banks operating in rural areas and insurance companies also retain veterinarians.
Another option is to work with drug researchers and manufacturing companies. Poultry feed and pesticide companies also appoint veterinarians. Wildlife parks, zoos, sanctuaries, zoological parks etc. need the services of veterinarians on a regular basis. Most of such establishments are managed by government bodies. Bulk milk producers and cooperative organizations need veterinary doctors.
Defence services and police departments need services of veterinarians to look after dogs engaged for security and investigation purposes.
There is good business opportunity for Veterinary doctors to pursue private practice by establishing their own clinic. Alternatively you may join a big clinic and offer your services as a member in doctors team.
Those interested in business and marketing may take up M.B.A. in Agri-Business after completing their B.V.Sc.
Teaching opportunities can be explored at the institutes listed above and others involved in Veterinary education.
Candidates with an aptitude for research may work as a research scientist at Indian Veterinary Research Institutes. Government of India has established a number of research establishments many of which conduct research on a particular animal. These organizations, as listed below, appoint candidates with higher qualification in veterinary science :
National Research Centre on Camel, Bikaner
Sheep Research Centre, Mannavanur, Tamilnadu
Central Institute for Research on Goat, Makhdum, Uttar Pradesh
Central Sheep & Wool Research Avikanagar, Rajasthan
Buffalo Research Centre, Hisar
National Research Centre on Meat, Uppal, Hyderabad
Central Research Institute on Cattle, Hisar
National Research Centre on Yak, Dirang, Arunanchal Pradesh
National Research Centre on Equines, Hisar
National Research Centre on Mithun, Jhampani, Nagaland
There are few other research Institutes dealing with animals health issues viz.
National Institute of Veterinary Epidemology, Bengaluru
National Institute of Animal Nutrition and Physiology, Bengaluru
CSIR-National Institute of High Security Animal Diseases, Bhopal
Scholarship based research opportunities can be explored in foreign countries.
Challenges
Animals also communicate but differently from humans. They can't explain what ails them so it requires extra efforts to understand their pains and problems and then decide on a course of action.
Skills and traits for success
Veterinarians must show compassion towards their animal patients and their owners. Willingness to learn, explore and to go the extra mile keep a veterinarian in good stead.
Useful websites to visit
If you want to further explore the field of veterinary and animal science, following websites of related organizations may provide you useful information:
SOURCE- EMPLOYMENT NEWS, volume-28, 12-18, October, 2019