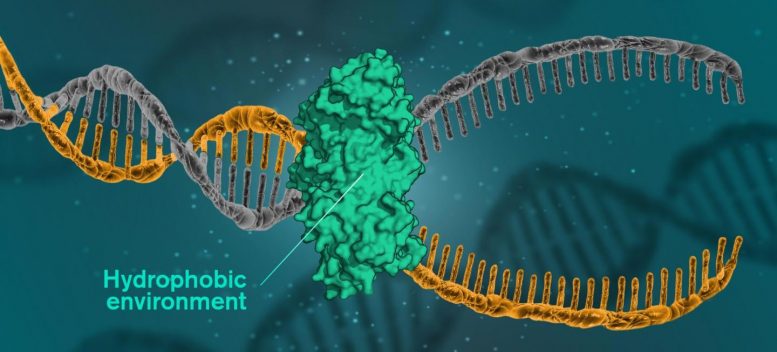
For
DNA to be read, replicated or repaired, DNA molecules must open
themselves. This happens when the cells use a catalytic protein to
create a hydrophobic environment around the molecule. Illustration
Credit: Yen Strandqvist/Chalmers University of Technology
DNA is constructed of two strands, consisting of sugar molecules and phosphate groups. Between these two strands are nitrogen bases, the compounds which make up organisms’ genes, with hydrogen bonds between them. Until now, it was commonly thought that those hydrogen bonds were what held the two strands together.
But now, researchers from Chalmers University of Technology show that the secret to DNA’s helical structure may be that the molecules have a hydrophobic interior, in an environment consisting mainly of water. The environment is therefore hydrophilic, while the DNA molecules’ nitrogen bases are hydrophobic, pushing away the surrounding water. When hydrophobic units are in a hydrophilic environment, they group together, to minimize their exposure to the water.
“We believe that the cell keeps its DNA in a water solution most of the time, but as soon as a cell wants to do something with its DNA, like read, copy or repair it, it exposes the DNA to a hydrophobic environment.” — Bobo FengThe role of the hydrogen bonds, which were previously seen as crucial to holding DNA helixes together, appear to be more to do with sorting the base pairs, so that they link together in the correct sequence.
The discovery is crucial for understanding DNA’s relationship with its environment.

Bobo
Feng, Postdoc, Chemistry and Chemical Engineering, Chalmers University
of Technology. Credit: Johan Bodell/Chalmers University of Technology
“We believe that the cell keeps its DNA in a water solution most of the time, but as soon as a cell wants to do something with its DNA, like read, copy or repair it, it exposes the DNA to a hydrophobic environment.”
Reproduction, for example, involves the base pairs dissolving from one another and opening up. Enzymes then copy both sides of the helix to create new DNA. When it comes to repairing damaged DNA, the damaged areas are subjected to a hydrophobic environment, to be replaced. A catalytic protein creates the hydrophobic environment. This type of protein is central to all DNA repairs, meaning it could be the key to fighting many serious sicknesses.
Understanding these proteins could yield many new insights into how we could, for example, fight resistant bacteria, or potentially even cure cancer. Bacteria use a protein called RecA to repair their DNA, and the researchers believe their results could provide new insight into how this process works – potentially offering methods for stopping it and thereby killing the bacteria.
In human cells, the protein Rad51 repairs DNA and fixes mutated DNA sequences, which otherwise could lead to cancer.
“To understand cancer, we need to understand how DNA repairs. To understand that, we first need to understand DNA itself,” says Bobo Feng. “So far, we have not, because we believed that hydrogen bonds were what held it together. Now, we have shown that instead it is the hydrophobic forces which lie behind it. We have also shown that DNA behaves totally differently in a hydrophobic environment. This could help us to understand DNA, and how it repairs. Nobody has previously placed DNA in a hydrophobic environment like this and studied how it behaves, so it’s not surprising that nobody has discovered this until now.”
More information on the methods the researchers used to show how DNA binds together:
The researchers studied how DNA behaves in an environment which is more hydrophobic than normal, a method they were the first to experiment with.
They used the hydrophobic solution polyethylene glycol (PEG), and step-by-step changed the DNA’s surroundings from the naturally hydrophilic environment to a hydrophobic one. They aimed to discover if there is a limit where DNA starts to lose its structure, when the DNA does not have a reason to bind, because the environment is no longer hydrophilic. The researchers observed that when the solution reached the borderline between hydrophilic and hydrophobic, the DNA molecules’ characteristic spiral form started to unravel.
“Nobody has previously placed DNA in a hydrophobic environment like this and studied how it behaves, so it’s not surprising that nobody has discovered this until now.” — Bobo FengUpon closer inspection, they observed that when the base pairs split from one another (due to external influence, or simply from random movements), holes are formed in the structure, allowing water to leak in. Because DNA wants to keep its interior dry, it presses together, with the base pairs coming together again to squeeze out the water. In a hydrophobic environment, this water is missing, so the holes stay in place.
Reference: “Hydrophobic catalysis and a potential biological role of DNA unstacking induced by environment effects” by Bobo Feng, Robert P. Sosa, Anna K. F. Mårtensson, Kai Jiang, Alex Tong, Kevin D. Dorfman, Masayuki Takahashi, Per Lincoln, Carlos J. Bustamante, Fredrik Westerlund and Bengt Nordén, 27 August 2019, Proceedings of the National Academy of Sciences.
DOI: 10.1073/pnas.1909122116













0 Comments:
Post a Comment